Literature
Determine the Molar Extinction Coefficient of protein in 3 small steps
November 15 2016, by Thomas Kofoed
Question:
How do I determine the molar extinction coefficient of my protein?
I have a purified protein that I would like to quantify accurately in my lab using 280 nm UV measurement (A280). To do so, I need to determine the molar extinction coefficient of the protein in my buffer system. What is the best way to do that?
Answer:
It is possible to experimentally determine the molar extinction coefficient (also known as the molar attenuation coefficient) of a protein. You do this by A280 measurements of a dilution series of the protein in known concentrations. A theoretical calculation can also predict an extinction coefficient. This is based on the number of A280 absorbing residues (Trp, Tyr, Cystine – disulfide bonds) [1].
However, the actual molar extinction coefficient depends on the buffer and 3-dimensional structure of the protein. Therefore, for accurate protein concentration determination by A280nm measurement, we recommend that you determine the extinction coefficient experimentally in the buffer you will use in your lab, for example, in PBS buffer [2].
Suggested three-step procedure to determine the extinction coefficient
To determine the extinction/attenuation coefficient, you will need access to an amino acid analyzer and a spectrophotometer. The analysis is then a three-step procedure:
- Measure the exact protein concentration
First, hydrolyze the sample in HCl in triplicate and measure the protein concentration by quantitative amino acid analysis (AAA), e.g., using a Biochrom 30+ instrument, which performs ion-exchange chromatography and post-column derivatization with Ninhydrin.
Avoid material in the buffer that can polymerize during the acidic hydrolysis, taking place at 110°C (e.g., sugars)Next, determine the molar concentration using the amino acid composition of the protein [1,2]. - Determine the UV absorbance
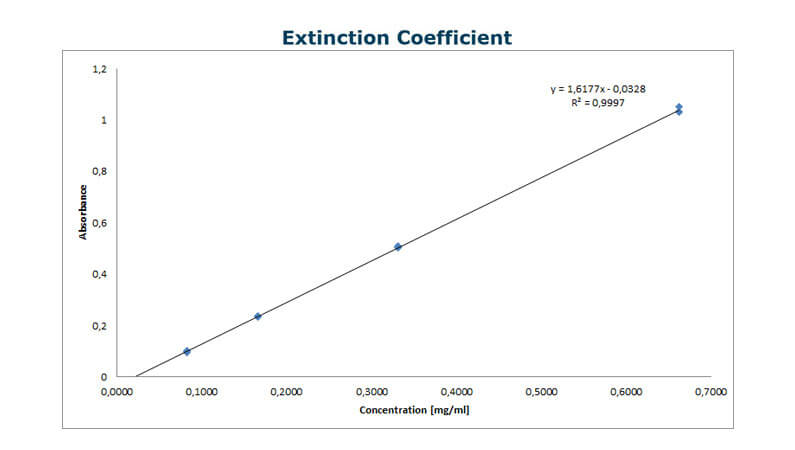
Based on the results from the AAA analysis, prepare a dilution series covering the highest possible concentration and a 50-fold dilution – and measure the UV absorbance at 280 nm.Now, derive a linear curve from the absorbance as a function of the concentration (see the example at the top of this page). - Calculate the molar extinction coefficient
Finally, using the Beer-Lambert law, calculate the absorptivity constant and molar extinction coefficient from the slope of the A280 curve and the molecular weight of the protein.

The analysis is typically within 10% accuracy. You can ensure this quality by running a BSA sample (NIST standard) along with your samples.
Things to consider when determining the extinction coefficient of proteins
You must determine the extinction coefficient in the buffer system you will use in your lab. Also, the sample should be as concentrated as possible. This ensures that you get an extinction coefficient covering as much concentration range as possible [1, 2].
UV measurement at 280 nm and triplicate amino acid analysis
If you do not want to experiment with this yourself, Alphalyse also offers an excellent service to help you determine the ext. coefficient of your protein.
Alphalyse combines UV measurement at 280 nm with accurate amino acid analysis. Find more information about Molar Extinction Coefficient determination on the Alphalyse website.
References
[1] Pace et al.: “How to measure and predict the molar absorption coefficient of a protein,” Protein Science, 1995
[2] Gill et al.: “Calculation of protein extinction coefficients from amino acid sequence data,” Analytical Biochemistry, 1989
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.