Literature
There is a terrific method for residual protein impurity detection, with detection limits down to low ppm levels. It can track and quantify the residual proteins through all purification steps and eventually the final biopharmaceutical product.
Before going into details with the method, let’s first look at the residual impurities often seen in process development – and why they increasingly concern the FDA and EMA.
The different types of residual impurities found in biologics
During the development and production of biologics such as cell & gene therapy products (CGTs), process-related impurities follow along with the product.
Typically, we divide impurities into three subtypes depending on their origin:
- Cell substrate-derived / product-related (e.g. host cell proteins, host cell DNA)
- Cell culture-derived (e.g. antibiotics, IPTG, DTT, growth factors)
- Downstream-derived / process-related impurities (e.g. enzymes, buffer components)
The last subtype consists of agents used to express and purify biological protein products. Standard process-related residuals include the Benzonase nuclease, enzymes for site-specific PEGylation, aminopeptidase, and Protein A. Other typical residuals are Tris, carriers, ligands, Tween/Polysorbate, DCA, TCEP, heavy metals, solvents, Triton-X, antifoaming agents, PEI, TFA/Acetate, Imidazole, etc.
Why are residual impurities of concern?
Investigating the levels of residuals is crucial, especially in the final product sample. Because the residuals can influence the stability and efficacy of the active ingredient, and may even pose a risk to the patient’s health [1-3].
With the development in sensitive analytical methods for residuals analysis, the FDA and EMA now seem more and more interested in data that map out the efficiency of the purification process for CGTs and vaccines [1-3].
How to fully document process-related impurity clearance
The FDA and EMA are asking an increasing number of CGT developers to document residual protein clearance before their biologic may into late-stage clinical trials [1-3].
The problem, however, is that traditional methods like ELISA and HPLC are restricted by low antibody-specificity and an insufficient limit of quantification, respectively. Since the residuals in samples are present at low-ppm levels, you need a highly sensitive and reproducible approach to get a detailed overview of the residual protein clearance [4, 5].
Fortunately, SWATH mass spectrometry (LC-MS/MS) offers precisely that. It fragments all peptides using Data Independent Acquisition (DIA). The method divides the defined mass range into small mass windows for MS/MS fragmentation of the peptides. Afterward, comparing the acquired MS data to an ion library of known peptides will identify the proteins [6].
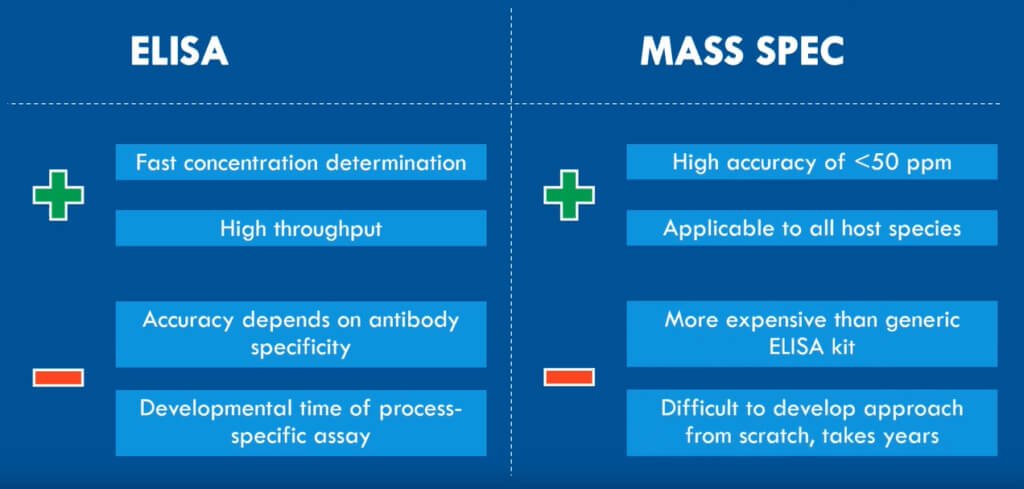
The advantages of mass spectrometry for residual protein analysis
Using SWATH LC-MS for analyzing residual proteins in CGTs include:
- Highly reproducible identification and quantification, due to the use of DIA mode
- Interference, from high amount of drug substance on the signal of low abundant residuals, is kept low by using small mass windows for MS/MS fragmentation
- Specific residuals can be followed throughout the entire process development. You can also use it to compare batches and for quality control after upscaling of batches
- A high throughput of samples makes it possible to quickly assess many individual samples from various steps in the purification
- A high sensitivity makes it possible to quantify low ppm levels
With these advantages, SWATH LC-MS is ideal for analyzing process-related residuals used in biologics development [6].
References
[1] U.S. Department of Health and Human Services – Food and Drug Administration: “Immunogenicity Testing of Therapeutic Protein Products — Developing and Validating Assays for Anti-Drug Antibody Detection: Guidance for Industry,” 2019
[2] U.S. Department of Health and Human Services – Food and Drug Administration: “Q3C Impurities – Residual Solvents: Guidance for Industry”, 1997
[3] European Medicines Agency: “Guideline on Immunogenicity assessment of therapeutic proteins,” 2017
[4] Zhu-Shimoni et al.: “Host Cell Protein Testing by ELISAs and the Use of Orthogonal Methods,” Biotechnology and Bioengineering, 2014
[5] Bracewell et al.: “The Future of Host Cell Protein (HCP) Identification During Process Development and Manufacturing Linked to a Risk-Based Management for Their Control,” Biotechnology and Bioengineering, 2015
[6] Heissel et al.: “Evaluation of spectral libraries and sample preparation for DIA-LC-MS analysis of host cell proteins: A case study of a bacterially expressed recombinant biopharmaceutical protein,” Protein Expression and Purification, 2018
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.