Case study
Revealing HCPs not detected by ELISA in antibody-drug product
Supplement ELISA with mass spectrometry data - and check for problematic HCPs
HCP ELISA and an initial mass spectrometry (LC-MS) analysis of our client’s monoclonal antibody (mAb) process samples indicated that it was very pure. None of the methods identified any Host Cell Proteins (HCPs) in 2 of 4 purification steps.
However, applying a semi-quantitative, native digest approach lowered the limit of detection (LOD) substantially, and the new analysis revealed more than 20 HCPs in the client’s final process step. We also used the data to evaluate the observed HCPs for potential problematic properties.
The client is a drug discovery company who develops therapeutics to treat cancer and autoimmune conditions. Their pipeline includes Fc fusion proteins, antibody-drug conjugates, and monoclonal and bispecific antibodies.
Usually, they apply commercial ELISA kits for HCP analysis. They were unsure if their ELISA reported all HCPs present, and they decided to try an orthogonal HCP analysis to look for specific, problematic impurities.
Step 1:
LC-MS assay using quantitative digest method showed no HCPs
The client sent us four samples from different purification steps to produce their mAb product expressed in E. coli. The primary aim was to get an overview of HCP clearance throughout the purification process.
Initially, we applied our quantification digestion protocol (the quantitative assay), a well-established method that works well for all sample types and expression systems. The analysis detected more than 20 HCPs in steps 1-2. Whereas analysis of steps 3 and 4 resulted in no HCPs above limit-of-detection (LOD).

Step 2:
Native digest substantially lowered limit of detection (LOD)
To check for HCPs below 1-5 ppm, we applied a semi-quantitative, novel digest method. This method digests HCPs for LC-MS detection while leaving mAbs largely intact.
Following the digest procedure, precipitation removed most of the undigested mAb. It can then increase the amounts of digested HCPs onto the LC-MS instrument, substantially expanding the assay’s sensitivity. As a result, we now detected over 200 HCPs in step 2. Interestingly, the analysis also found more than 20 HCPs in steps 3 and 4.
With the results, the client could compare the results to their ELISA estimations. Furthermore, it was possible to look for HCPs that may be problematic for product stability and patient safety – even below one ppm.
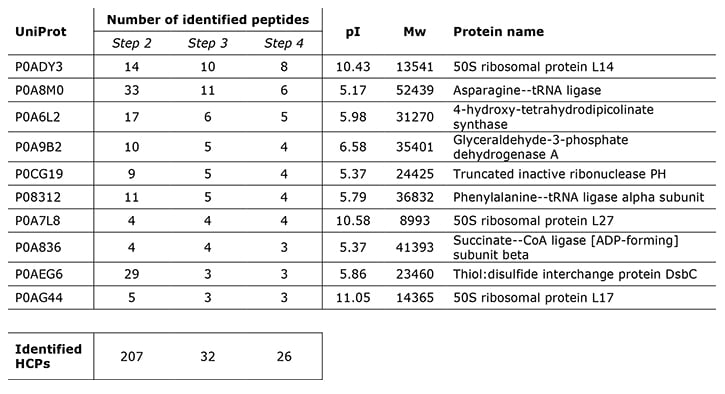
Top 10 HCPs identified in the process steps 2-4 with the semi-quantitative, native digest method. Instead of ppm, the table shows the number of detected peptides originating from the proteins.
Talk to us
Whatever protein-related challenge or question you may have, we would love to help. Our experts can help you decide on the best analytical approach for your project by email or online meeting - providing advice without obligation.